NIDM Apps
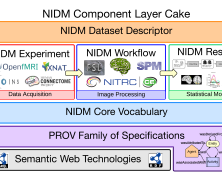
The Neuroimaging Data Model (NIDM) is a general framework for data exchange that is defined using a set of community developed specifications to convey neuroimagingdata and meta-data, developed by the INCF Neuroimaging Task Force.
Summary
The Neuroimaging Data Model (NIDM) is a general framework for data exchange that is defined using a set of community developed specifications (http://nidm.nidash.org/specs/) to convey neuroimagingdata and meta-data, developed by the INCF Neuroimaging Task Force (http://wiki.incf.org/mediawiki/index.php/Neuroimaging_Task_Force).
The ultimate scope of NIDM is all types of neuroimaging data, encompassing provenance from raw data to final statistical inference, but is presently focusing on fMRI and representation of the final statistical results. Export using NIDM is now implemented natively in SPM12 and available in FSL. We would like to invite Hackathon participants to build applications using NIDM. We will make available a set of fMRI experimental results exported using NIDM. Possible applications include (but are not limited to):
- Create ready-for-publication tables (PDF/HTML/RTF?) from peak data in NIDM.
- Create a machine-generated paragraph describing the model of each study, ready for inclusion in a manuscript, using the model information in NIDM.
- Create a conversion tool, exporting peak data for coordinate-based meta-analysis tools (such as GingerALE http://www.brainmap.org/ale/, MKDAhttp://wagerlab.colorado.edu/files/tools/meta-analysis.html, or SDM, http://www.sdmproject.com/)
- Create a JavaScript viewer for NIDM results using existing components such as Papaya viewer (e.g.: https://github.com/incf-nidash/nidmviewer)
- Make existing utilities (such as nib-ls from https://github.com/nipy/nibabel/ NIDM aware).
Contact
Tom Nichols for NIDASH
Neuroimaging-TF@incf.org
http://nidm.nidash.org/specs
